SPICA: Surface Property fItting Coarse grAined model
Tutorialwith MD software
Top
Several different MD softwares are useful for MD simulations using the SPICA force field:
LAMMPS is the first choice for the SPICA force field.
GROMACS might show a better performance,
though a patch file is required to make it useful.
MPDyn2
was originally used to build the previous SDK force field,
even though a good performance on parallel computers is not expected.
MPDyn2 could be useful for analyses.
In this tutorial, we focus on LAMMPS and explain how to prepare input files for
different systems.
Prerequisites
We will use a command-line tool, cg_spica, to generate some input files required to
run coarse-grained MD using SPICA.
The tool's repository is open on GitHub and found at
here.
README in the repository mentions required enviroments and installation to use the tool.
Please make sure that you can excute the command-line tool on a console before starting this
tutorial.
Contents
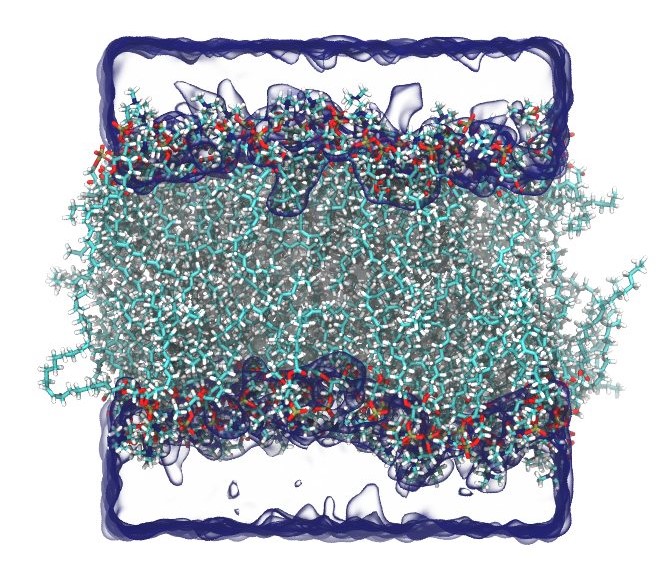 Lipid membrane |
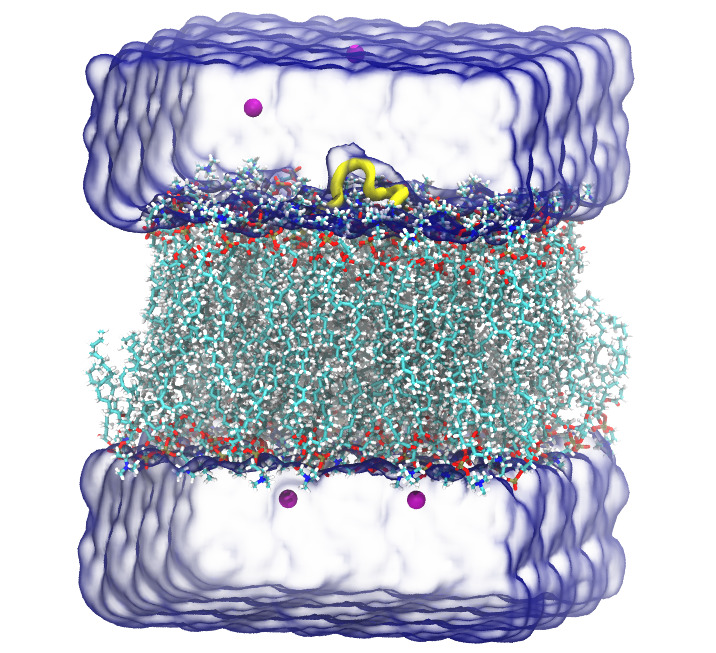 Protein |
|
|
|
SPICA Force Field
Okayama University
